Data simulation for linear models in R
Als Kalendereintrag speichernCourse content information subject to change
OUTLINE/AIMS/CONTENT: Usually, when we fit linear models, we don't know the 'truth' about the data we analyse, and hope that the model uncovers this 'truth' (or at least reveals something related to it). However, this is not necessarily the case. The examples probably most well-known are lack of power (i.e., a model doesn't reveal significance despite the effect in question actually existing in reality) and type I errors (i.e., the model erroneously reveals an effect which actually does not exist). More generally, for each model we fit, we have to face the possibility that the results reflect the processes investigated only to some extent, but in parts also reflect problems with the model/the data. This becomes even more of an issue when the models used are recent developments with, in part, little known properties (such as GLMMs) and/or when the combination of model complexity and sample size pushes against the limits of what models can do (which is what quite frequently happens).
The primary aim of this course is to teach how to simulate data in a framework of linear models and using R. The cool thing about simulated data is that one knows exactly what is 'true' (because one has generated the data). Hence, one can then compare what a model reveals with truth. The perhaps most well-known use of simulated data is 'power analysis' (determining the probability of an analysis to reveal significance, given a certain effect and sample size, an alpha level, and a test). However, it can also be used to address questions such as 'Does the method do what I think it does?', 'Does the method do what people think it does?', 'Does bias in sampling translate into bias in the estimates?', 'How does an analysis behave when its assumptions are violated?', 'What is the probability that the model converges at all?', 'What is the expected width of confidence intervals', 'What is the precision of the estimated coefficients?', and many others.
The course has three main aims (in order of increasing importance):
- teaching how to simulate data and conduct power analyses in a wider sense (i.e., addressing the kinds of questions mentioned above). As part of this, participants will, for instance, learn how to simulate data sets with respect to various predictors (e.g., crossed and nested factors, correlating covariates), different response variables (e.g., Gaussian, binary, Poisson, negative binomial, beta, etc.), effects of fixed effects predictors and grouping variables (i.e., random intercepts and slopes), and non-linear effects of covariates. Participants will also get to know various functions for random sampling.
- teaching a bit about programming in R. In fact, it is somewhat in the nature of simulations that more or less the same set of operations needs to be done many times. In the course, participants will learn how to automate i) processes in R (using for- and while-loops and also lapply), ii) write code that evaluates conditions (e.g., if-constructs), iii) speed things up where possible, iv) store results, v) write one's own functions, vi) catch and deal with errors and warnings, vii) parallelize code execution, etc.; nothing really fancy, though, just the basics needed to program such simulations.
- enhancing the understanding of linear models and what the link between them and 'life' (in either direction) is. In fact, simulating data means that one needs clear hypotheses about life and an equally clear understanding of how these hypotheses translate into model coefficients (and eventually a response). So in case you are not quite sure whether you really understand the precise meaning of such things like interactions, link functions, error distributions, random intercepts and slopes, or what the idea behind an offset term is, the course might be useful for you.
The course will closely follow the course ’Linear models and their application in R’ (course number 34097) as we shall begin with simulating simple linear regression, and step up to simulating (effects of) factors, interactions, non-linear effects, data for GLMs (i.e., non-Gaussian response variables), and finally effects of grouping variables (i.e., random intercepts and slopes).
STRUCTURE: The course will consist of lectures/seminars, practical applications/examples, and extensive exercises. The exercises will by mandatory and an integral part of the course. That is, taking the course without working on the exercises is not an option as much of the material to be learned is put into the exercises. The exercises will take place during the regular course time (see below), and I'll be available in case participants encounter insurmountable obstacles.
TARGETED AUDIENCE: The targeted audience is not limited to students, but also faculty members, post docs, etc. are welcome to attend (basically everyone interested in the topics of the course).
REQUIREMENTS: The course requires a fairly good grasp on linear models as I teach them in the linear models course. Hence, participation is only permitted for people who participated in the course ’Linear models and their application in R’, either the most recent instance (GGNB course number 34097 in the summer semester 2021) or previous rounds. In addition, participants need to be familiar with the basics of R, also bring some level of interest in programming (but knowledge of a proper programming language is not required), and they shouldn't be too shy of a little bit of math. An additional requirement that needs extra time during the course is that, in preparation for each day, participants will need to go through a few handouts of the linear models course again.
MATERIAL PROVIDED: The course is accompanied with plenty of handouts which will be made available during it.
WHEN & WHERE: The course will take place during the three weeks from Nov. 1st to Nov. 18th, 2021, with lessons taking place each weekday from 9:00 (sharp) to at latest 17:00 (with several shorter and one longer break). Venue is the German Primate Center, Kellnerweg 4, Seminar room E0.14. The minimum number of participants is 12, the maximum number is 20. .
CREDITS: Certificates of participation are accepted to fulfil credit requirements of the PhD programme Behavior and Cognition as well as the GGNB programmes (for GGNB, 4C are given). A regular attendance is required. Students of other PhD programmes should inquire with the heads of their study programmes regarding credits.
LANGUAGE: the course language will be English.
SIGNING UP: by sending an email to statistics_teaching(at)dpz.eu; when registering, include your full name, affiliation, status (faculty member/PostDoc/PhD, Master) and study programme (if applicable). Please confirm that you have participated in the workshop “Linear models and their application in R” before and include when you participated in that workshop. You can register between September 19 and October 7, 2022. People signing up later cannot be considered. Due to the priority regulations (see below), signing up does not imply that you can participate. Everyone who signed up will receive an email soon after October 7th, stating whether s/he can participate.
BEFORE YOU SIGN UP: please be aware that the course requires a more or less full time commitment for three entire weeks. Please sign up only if you will be definitely able to participate (given no major unexpected events). 'No shows' and late sign-outs will have reduced priority of access in future rounds of the course; GGNB and GAUSS doctoral students will be banned from all skills and methods courses and funding options for 12 months.
PRIORITY OF ACCESS: there is a limited number of slots available, and in case more people sign up for the course than can participate, priority of access is given to first, members of the Leibniz ScienceCampus Primate Cognition, second, staff, members, and students of the University of Göttingen and the German Primate Center, and finally (third), to everyone else. Within each group, priority of access is given in the order in which people have signed up. Finally, in future rounds of the course people that have signed up for this round but then signed out again or just didn't appear might get lower priority.
Referent/-in
Dr. Roger Mundry
Anfahrtswege zum DPZ
Lageplan des DPZ
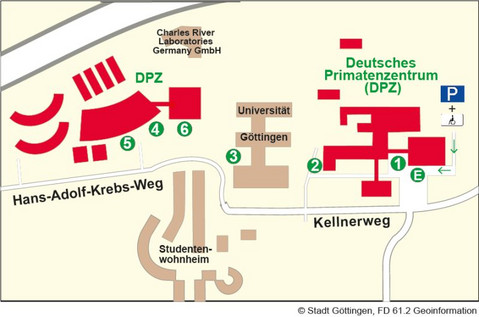
E - Haupteingang/Anmeldung
1 - Geschäftsführung; Abteilungen: Infektionsbiologie/-modelle, Versuchstierkunde, Primatengenetik, Verhaltensökologie und Soziobiologie, Kognitive Ethologie, Neurobiologie; Verwaltung; Bibliothek; Stabsstellen: Forschungskoordination, Kommunikation, Informationstechnologie, Betriebstechnik
2 - Materialanlieferung/Einkauf
3 - Forschungsplattform Degenerative Erkrankungen; Forschungsgruppe Soziale Evolution der Primaten
4 - Abteilung Kognitive Neurowissenschaften
5 - Tierhaltung
6 - Bildgebungszentrum; Abteilung Funktionelle Bildgebung
Anreise mit dem PKW
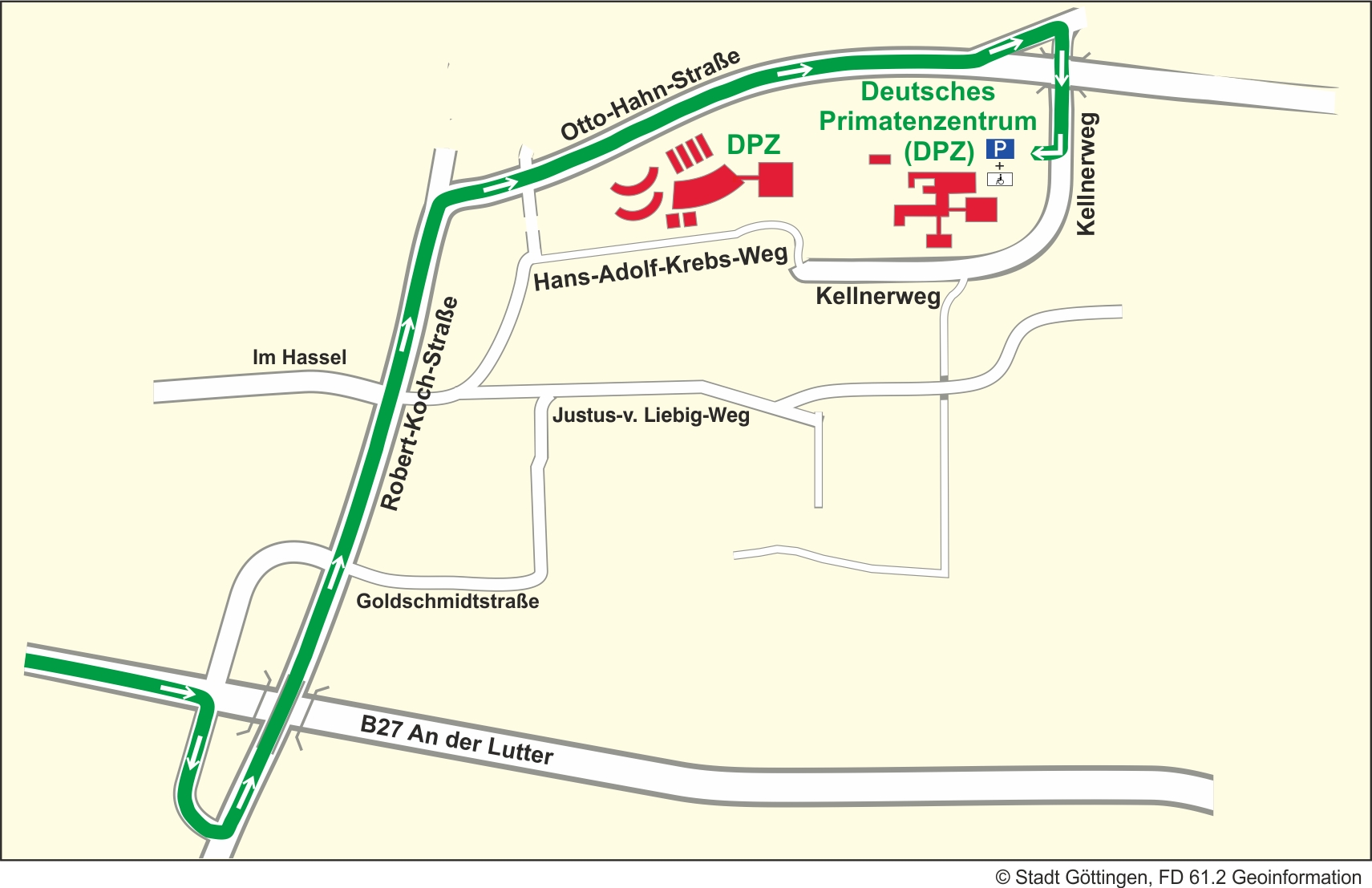
Folgen Sie von der Autobahnausfahrt "Göttingen Nord" der B27 in Richtung Braunlage bis zur dritten Ampelkreuzung. Biegen Sie rechts ab Richtung Kliniken und anschließend links in die Robert-Koch-Straße. Am Ende der Straße fahren Sie rechts in Richtung Nikolausberg auf die Otto-Hahn-Straße. Die erste Straße zu Ihrer Linken ist der Kellnerweg, das Primatenzentrum ist ausgeschildert.
Anreise mit dem Bus
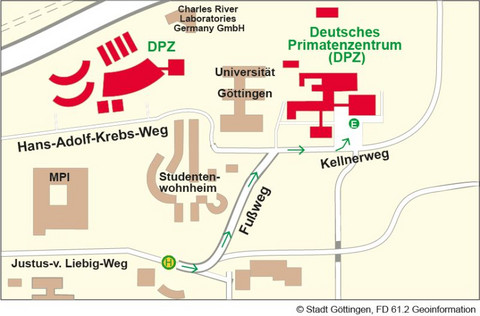
Ihr Fußweg von der Bushaltestelle Kellnerweg zum DPZ-Haupteingang/zur Anmeldung:
Von der Bushaltestelle Kellnerweg (Linie 21/22 und 23) Straße überqueren, in Fahrtrichtung des Busses gehen. Am Briefkasen links in den Fußweg einbiegen und rechts halten. Am Ende des Fußwegs rechts in den Kellnerweg abbiegen. Der Haupteingang des DPZ liegt dann auf der linken Seite.
Datum und Uhrzeit 01.11.22 - 09:00 - 18.11.22 - 17:00 Auf max. 20 Teilnehmer begrenzt. Anmeldung erforderlich
Veranstaltungsort Seminar room E0.14, German Primate Center, Kellnerweg 4
Leibniz ScienceCampus Primate Cognition
statistics_teaching@dpz.eu
